How DNA Sequencing Works: Sanger and NGS
Idea creds @Infusiondx
but first, quby:
DNA sequencing is a valuable biotechnique used for disease diagnosis, genetic testing, evolutionary biology, and more. One of the earliest methods of sequencing, Sanger sequencing, also called dideoxy chain termination utilizes concepts of cellular DNA replication to deliver its results.
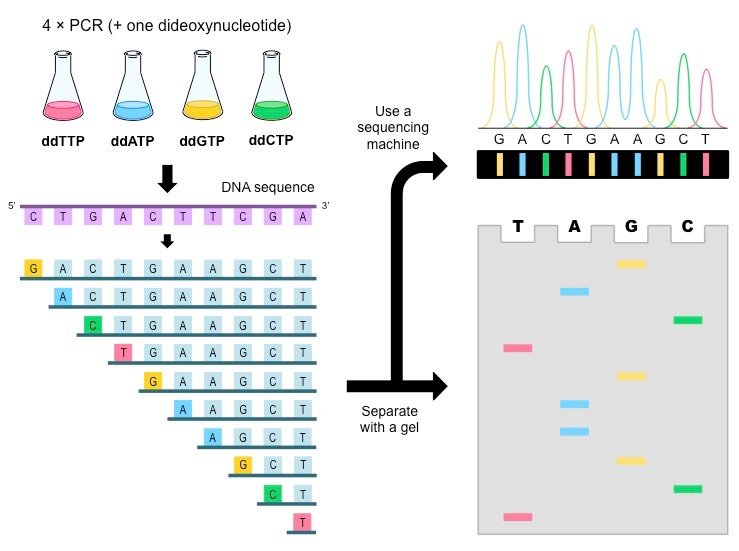
The process for Sanger sequencing is as follows. First, the strand of DNA to be sequenced is placed in four different tubes. To each tube, we add a solution of all 4 deoxynucleotide triphosphates (dNTPs): deoxyadenosine triphosphate (dATP), deoxyguanosine triphosphate (dGTP), deoxycytidine triphosphate (dCTP), and deoxythymidine triphosphate (dTTP). These 4 dNTPs are very simple: each dNTP = deoxyribose (ribose sugar without an oxygen) + nitrogenous base (adenine, guanine, cytosine, or thymine) + 3 phosphate groups (Pi). (Notice that dATP is basically ATP, adenosine triphosphate, but without an oxygen!)
Each tube now has the DNA to sequence + dATP + dGTP + dCTP + dTTP. Each tube now gets one different dideoxynucleotide triphosphate (ddNTP). These are basically dNTPs, but with another oxygen removed. ddNTPs are special because they do not allow extension of the DNA chain after they've been added. Why? Let's look at the chemistry involved.
Here is dNTP vs. ddNTP:
Here is a double-stranded DNA chain:
Here is how dNTPs are normally added to the DNA chain:
Look at the yellow box. Do you see the 3' hydroxyl (OH) group sticking out the bottom of the base of the growing chain? The OH is trying to connect with the oxygen of the phosphate group. This is how elongation works if a dNTP was just added.
However, imagine that the black DNA chain had a ddNTP instead. The 3' OH sticking out would instead be a 3' H, which doesn't allow this connection. So, the effect of adding a ddNTP is that the chain can no longer be grown after addition. (Now think about why Sanger is also called dideoxy chain termination.)
Let's go back to the Sanger process. Here are the contents of our 4 tubes:
1 = ddATP + DNA to sequence + dATP + dGTP + dCTP + dTTP
2 = ddGTP + DNA to sequence + dATP + dGTP + dCTP + dTTP
3 = ddCTP + DNA to sequence + dATP + dGTP + dCTP + dTTP
4 = ddTTP + DNA to sequence + dATP + dGTP + dCTP + dTTP
Also note that all ddNTPs are fluorescently labeled for easy detection later on.
To kickstart our process, we add DNA polymerase and primer, which help add the d/ddNTPs to the template DNA and help anchor the DNA pol + get it started, respectively. We let the DNA polymerase do its thing. What we end up with are many partially-replicated DNA strands with fluorescently-labeled ddNTP stop points at different locations. Because replication progress varies, each DNA strand will have a different mass due to different amounts of nucleotides being added. Because each DNA strand has a different mass, we can use gel electrophoresis (GE) to separate the DNA!
Here's a super basic explanation of GE: https://www.youtube.com/watch?v=ZDZUAleWX78&vl=en. In short, the longest strands stay at the top and the shortest ones migrate to the bottom. We do GE in 4 columns, one for each ddNTP type (although we can do it in one column, but we would need to color each ddNTP in a different color for this to work). We get a gel that looks like this:
The sequence of the complementary strand is read from bottom to top - the shortest strands go to the bottom, so this would be the beginning of the strand. Important tip to solve Sanger problems: the sequence of the original template strand is NOT the sequence on the gel. It is the complement of the sequence on the gel, read from the bottom up.
There's newer versions of Sanger like Next Generation Sequencing (NGS) which are basically automated versions of Sanger that can crunch samples WAY faster.
So yeah. Here are some additional resources to learn more, and leave a comment/DM me if you have questions/comments - thanks!