DNA Repair Mechanisms You Need to Know for the USABO
Idea creds @hyun.hooe.
also listen to EVILJ0RDAN. keep streaming he’ll drop another trust
DNA is fundamental to cellular function because it provides the blueprint for protein creation. Cells have many mechanisms to reverse DNA damage.
There are two broad groups of repair mechanisms: those that fix local disruptions (direct repair, base excision repair, nucleotide excision repair, and mismatch repair) and those that fix double strand breaks (homology-directed repair and non-homologous end joining).
Category 1: fixing local disruptions
Direct repair is how thymine dimers and chemically modified bases are fixed.
Thymine dimers result from UV damage, and make the DNA bulky, ruining the DNA replication process and contributing to carcinogenesis. To fix thymine dimers, an enzyme called photolyase breaks the bond between the two thymines in a process called photoreactivation.
Radiation, oxidative stress, certain chemicals, and other environmental factors can cause faulty base modifications, including methylation when not needed. Alkyltransferase removes alkyl groups (formula: −CₙH₂ₙ₊₁, which includes methyl (CH3, n=1)) to restore bases back to their natural form.
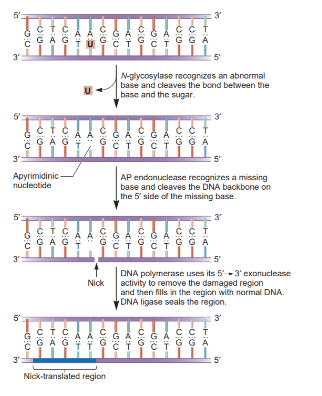
Base excision repair (BER) repairs chemically modified bases using DNA glycosylases, which excise the modified base, leaving an empty space called an abasic (AP) site. Then, AP endonuclease cleaves the backbone in that region to create accessible 3’ hydroxyl and 5’ phosphate sites. DNA polymerase I and DNA ligase then fill this gap. (This is the E. coli simplified version, btw.)
Nucleotide excision repair (NER) repairs bulkier modified bases (like thymine dimers for which photolyase repair wasn’t enough) and other big problems. Four proteins are involved in NER in E. coli: UvrA, UvrB, UvrC, and UvrD (a helicase). First, the UvrA-UvrB complex scans for and identifies the site of the helix distortion. Upon identification, UvrA dissociates and UvrC binds UvrB, still at the site. The UvrB-UvrC complex cleaves around the damaged site, and UvrD unwinds this area to remove the cut site. DNA polymerase I and DNA ligase then fill this gap. Deficiencies in NER actors most notably result in xeroderma pigmentosum (XP), characterized by a high sensitivity to UV light.
Mismatch repair fixes mismatched bases in DNA caused by faulty DNA polymerase III activity during replication. The repair system in E. coli is as follows. A MutS dimer identifies the site of mismatch external to the helix. A MutL dimer finds and binds the MutS dimer. MutH (an endonuclease) binds the MutS-MutL complex, and is activated by MutL. MutH searches for the nearest GATC sequence with a methylated adenine and uses that as one of its cutting points on the daughter strand (it’s just a specific thing it chooses to cut at, much like a restriction enzyme). UvrD helicase unwinds the daughter strand until a bit after the mismatch site, and exonuclease removes this entire area. DNA polymerase I and DNA ligase then fill this gap.
You may ask: how does MutH know which strand is the daughter? How does it choose what to cut? Don’t both strands look the same? Which one is truly the mismatched one, and which is the original correct one? Note that mismatch repair often occurs soon after DNA polymerase III synthesizes the daughter strand, so epigenetic patterns (methylation, etc) have not yet been established. As a result, mismatch repair occurs on hemimethylated DNA, which has a methylated parent and unmethylated daughter strand. This is how MutH knows.
Category 2: fixing double strand breaks
Here’s a very basic explanation of homology-directed repair (HDR) and non-homologous end joining (NHEJ).
HDR: The ends of the DNA in the double strand break are processed, which means some extra cuts are made so the ends are jagged and no longer flat. This helps create a template in the beginning portion (kind of functioning like a primer) so that extending the strand is a bit easier. The homologous chromosome’s DNA strand has the same sequence as this one, so it’s used to determine how to extend the strand for places that have been processed (cut off) on both strands. As always, pol and ligase stitch everything back up.
NHEJ: A little bit of processing occurs around the site of the double strand break. Then it’s just stuck together with pol and ligase, and some proteins hold the pieces together. Pretty haphazard, and very error-prone. But when there’s no homolog, we have no reference, so we can’t do much better than this.
Here’s some more stuff.
Here’s a cool application of HDR and NHEJ in a different context: https://www.sixfootscience.com/brain-snips/it-doesnt-get-crispier-than-crispr-an-overview-of-the-revolutionary-gene-editing-technology
Mismatch repair: https://youtu.be/wukucowb8sA?si=7fVZLq0QyhzPWYTV
All images from Brooker’s - W book.